Artificial intelligence (AI) has recently become a popular term in the technological world. AI refers to the simulation of human intelligence with the capacity for achieving goals within computers. Machine learning (ML) – a subtype of AI – refers to a statistical model that is able to independently learn to make inferences on new data based on data it has previously analysed. ML has markedly improved efficiency in multiple analytic domains including voice recognition, handwriting recognition, targeted marketing and robotics.1 ML is a broad term that includes a family of methods extending from decision tree models to neural networks. Each method has different attributes that relate to their suitability for a given task. Interestingly, despite their recent popularity, neural networks have been in existence since at least the 1950s, when Marvin Minsky used an artificial neural network to solve a maze.2 However, only in recent times has computing power sufficiently improved to allow the wider application of computationally intensive ML methods.
Aside from computing power, the other major development extending the reach of ML has been data acquisition and storage – an essential component to ML. Big data is a term used to describe the increasingly large and more complex datasets that form the basis for ML models.
Modern times have seen increasing accessibility and use of large volumes of data. This is apparent in medicine with the advent of electronic health records (EHRs), implantable electronic devices and advanced wearable monitors, all of which record unprecedentedly large volumes of biological data every second.3 ML has already contributed to healthcare advances in a number of specialties. This has taken various forms from assisting imaging and pathological diagnoses, to identifying novel disease risk factor associations and phenotypic subgroups.4,5 Rajpurkar et al. used a convolutional neural network to develop a model that detected 14 different pathologies from a total of 420 frontal chest radiographs at a much faster rate than board-certified radiologists (1.5 minutes versus 240 minutes for all 420 radiographs). Their model was trained from a database of over 100,000 chest radiographs and could also anatomically localise where the pathologies (including pneumonia, effusions, masses and nodules) were present.6 As an example of unsupervised learning, Horiuchi et al. identified three phenotypic groups of acute heart failure patients that significantly predict mortality and re-hospitalisation.7 Within cardiac electrophysiology, near-term applications of novel data sources have begun appearing. Many of these are driven by industry–academic–healthcare partnerships, the most famous example of which is the Apple Heart study.8 However, significant late stage and outcomes data remain to be seen.
These early studies demonstrate some applications of ML in healthcare.9 Updating prior understanding and clinical practice developed over years will take time, but ML has the potential to improve disease definitions, classification and management.
Big Data
Data are a core requirement of ML methods. Theoretically, ML methods can be applied to datasets of any size. However, ML methods often benefit from large datasets as they provide more experience on which to train a model. ‘Features’ are components of a dataset that describe characteristics of the observations being studied. These features are fed into computational models that can then provide insight into the observations, for example clustering of similar observations into groups or prediction of outcomes. Large datasets used in this context are referred to collectively as big data. There are many sources of big data in modern healthcare including EHRs, biobanks, clinical trial data and imaging datasets. The advent of ML methods has allowed enhanced insight into these resources.
The Three Vs of Big Data
There are three attributes that delineate the attributes of big data. The first V refers to volume of data. The larger the volume, the more features and experience a model can be trained and tested on. The second V refers to the velocity of data, which describes the speed at which the data is generated. With a higher velocity of data, models can remain more clinically relevant as they are retrained on current experience. The third V refers to variety, which reflects the diversity of the types of data contained in a dataset. Diverse data allows more features that can, in turn, increase generalisability and potentially the accuracy of the model.10 As a general rule, an increase in any of these attributes create a larger demand on the hardware and complexity of the method used to generate a model.
Potential Pitfalls of Big Data
The issues with big data can be related to the abovementioned Vs. High volume and velocity data requires significant computer processing power to analyse. Typically it is not an isolated volume or velocity issue, but a combination of the two together. This need for computational efficiency is one of the reasons novel ML methods outperform traditional statistical methods.
The extent of resources required to store and analyse data can be prohibitive. This limits the translation of data into clinical investigation and clinical practice. This is relevant in electrophysiology where cardiac devices create large volumes of data each second.
Another issue with big data is that it is often poorly organised and managed. This problem, also observed in small datasets, is amplified on a larger scale. Inaccurate data can be inappropriately included or not recognised for its limitations. Thus, the problem of inaccurate data producing inaccurate models remains.
Data sharing is an increasingly popular concept made easier with improvements in technological infrastructure.11–13 Much of this is currently used for subgroup analyses, validation of prior work or the exploration of new hypotheses using trial data. Sharing of data will allow future researchers larger volumes of data to train and test models on. Many models exist for effective data sharing. Federated database systems allow geographically separate databases to be connected via networks. Thus, without merging the databases, researchers can submit queries to the federated database that interfaces with each individual database and provides results from all of them. This method allows the individual databases to remain heterogeneous and distinct. In contrast to this, distributed learning is a method that allows an ML algorithm to be trained using separate datasets. A central ML model is created and updated based on the training performed in each dataset. This model allows maintenance of data security and privacy whilst still harnessing the size of multiple datasets. However, opinions on data sharing as regards privacy and intellectual property, particularly in the medical industry sector, may remain a barrier to the spread of data sharing.14
Machine Learning Methods
ML algorithms use and require significant datasets in order to create models and test them. The foundation of ML is creation of a model on a training dataset with subsequent validation of that model on an evaluation dataset (both of which are often subsets of an original large database). The model is then often run on a test dataset to provide an unbiased evaluation of its performance. The training dataset is used to fit the weights of a model, which detail the relationship between inputs and outcomes in a way specific to the chosen model. The validation dataset is then used to fine tune the model hyperparameters and evaluate the model. Once validated, models can be improved by being re-trained on new data. After re-training, models need to be validated again. These can increase the generalisability of the model across different populations/datasets. This allows the on-going improvement of the model with rapid responses to changing epidemiological or clinical patterns.
Machine Learning Computational Approaches/Algorithmic Principles
The overarching principle of ML is the use of training, evaluation and test datasets to create a valid model.15 ML methods are broadly categorised as supervised, unsupervised or reinforcement learning. Reinforcement learning aims to refine a strategy in a controlled environment stochastically. Reinforcement learning is outside the scope of this article.
Knowing which machine learning technique to apply is essential to achieving the objective, analogous to choosing an appropriate statistical technique in traditional methods or choosing an appropriate study design in epidemiology.
Supervised Learning Methods
Supervised learning (SL) algorithms aim to classify input data to the correct outputs based on prior input-output pairs that are correctly labelled. The need for labelling can be time consuming. However, the methods are very effective at classification using large datasets.
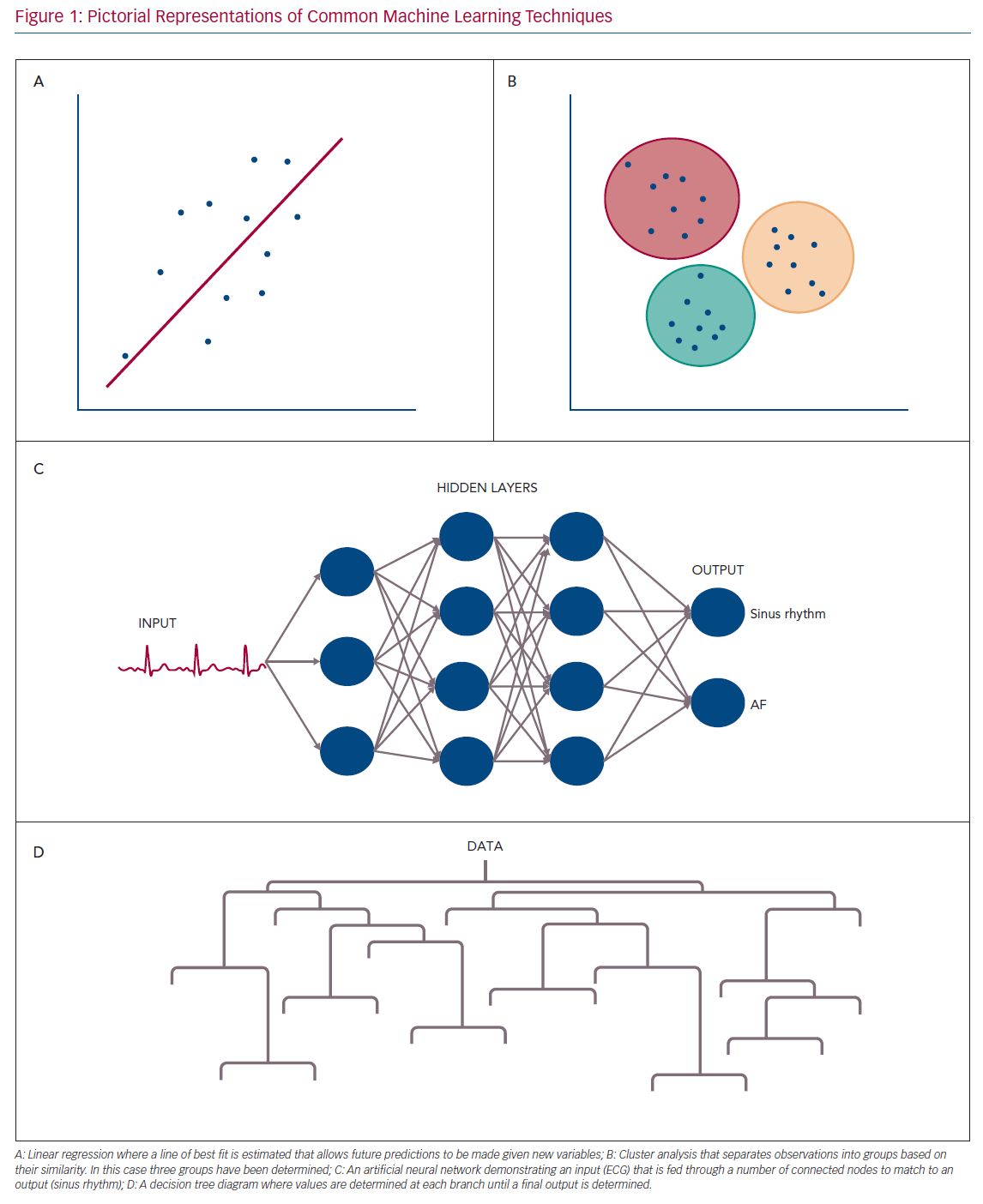
Examples of ML techniques used in SL include traditional linear and logistic regression, artificial neural networks (ANNs), support vector machines (SVMs), decision trees and random forests (RFs).15,16 Pictorial representations of common ML approaches are shown in Figure 1.
ANNs and SVMs are among the most common SL methods. They require intensive computational power and time and are prone to over fitting (where the model fits well for the training data but poorly for any new data). However, they are flexible regarding assumptions about the data and offer significant improvements in large datasets where traditional statistical methods struggle.17 ANNs can be layered and altered in order to offer more efficiency in dealing with complex and large data.
Decision trees and RFs are closely related. Decision trees are useful with smaller and simpler datasets. They classify data in a binary fashion along a chain of steps. Each branch following a step then has further classification at the next level. This then spreads out until an outcome is reached based on the series of steps taken to get there. The pictorial representation gives the shape to which decision trees are named.
RFs are an extrapolation of decision trees where multiple decision trees are combined and each tree is independently trained and verified. They also have the ability to exclude individual trees or components leaving them robust to outliers in meta-analysis and less prone to selection bias.
SL has its limitations including over fitting, the requirement of training and validation and the requirement for accurately labelled data, which can be laborious.16 SL models can have poor generalisability and struggle transferring experiences to new data owing to over fitting whereby the model is very well fit only for the original dataset. As a result different models are often required for data from different populations. Each model requires large volumes of labelled data to train and then validate. Importantly, SL does not necessarily overcome prior human biases in classification owing to the labelling step, which is often performed by humans and thus translates those biases into the model. This has had important implications in algorithms overweighting the importance of correlations as causation. This was the case with the Correctional Offender Management Profiling for Alternative Sanctions tool that analysed recidivism and was found to be using race as a predictor in its algorithm.18 Each of these limitations result in varying degrees of bias, impacting on the data and future results.
Unsupervised Learning Methods
Unsupervised learning (UL) has a different goal to SL. In this method, the algorithm attempts to find patterns in the data without prior labelling. For example, in clustering the algorithm creates an unbiased set of categories without human intervention based on similarities between observations (e.g. creating groups of people based on their similarities). There are a variety of techniques included in UL, such as clustering, autoencoders and principal component analysis.15–17
UL is particularly useful when the dataset is very complex or if there is no natural fit to the data. In medicine, it is increasingly being used to identify disease phenotypes in heterogeneous conditions. This could suggest new classifications or stratifications that lead to more sophisticated treatment allocations. It can also be used in less established diseases, where data or literature are poorly developed or pathophysiology is not completely understood. As an example, Shah et al. clustered heart failure with preserved ejection fraction into phenotypic subtypes based on cardio-metabolic, cardio-renal and biochemical features.19
UL can produce multiple cluster schemes after analysis. Choosing which cluster scheme is most accurate or appropriate can be difficult and requires clinical interpretation. Importantly, just because a model defines clusters within a disease does not mean these clusters have any clinical relevance. For this to be successful sometimes it is necessary for UL to be combined with follow-up studies assessing the relevance and utility of these clusters. For example, Ahmad et al. identified differing mortality risks after clustering groups of patients with chronic systolic heart failure.20
Machine Learning Applications in Cardiac Electrophysiology
ML is gradually expanding its utility in medicine, particularly in cardiology. Cardiac electrophysiology is particularly suitable for ML methods given its big data use and need for more accurate disease phenotype definitions and risk prediction. Returning to the three Vs, through implantable electronic devices, intracardiac mapping and wearable devices, high volume, velocity and variety data are produced. ML methods provide an opportunity to extract maximum clinical and public health benefit from this data.
Surface Electrocardiography
Surface ECGs can provide non-invasive, cheap and detailed information regarding arrhythmias. Importantly, because of these attributes, improving ECG interpretation could have substantial public health benefits.
There is a bulk of research involving ML and ECGs. While computer analysis of ECGs has been available on most machines for many years, these are often inaccurate because of the quality of ECG recorded. Traditional methods have relied on examining R-R intervals and P-wave presence. However, newer ML-based models have undertaken more sophisticated feature extraction to analyse a variety of rhythms.21
Research on ML in ECG began with basic improvements in pre-processing such as noise reduction or extraction of features such as P-wave or QRS complex characteristics.22,23 This initial work laid the foundation for future research to use these algorithms to create features that could be fed into classifiers such as ANNs and SVMs to further analyse ECGs. Much of the work performed in this area uses the MIT-BIH publicly available classified data that consists of 25 ECG recordings that are 10 hours long.24–26 Importantly, this should herald caution in the assessment of diagnostic accuracy in such studies given the small sample sizes.
Many investigators have attempted to optimise the detection of AF using differing methods, duration of recordings and leads. Models differ by analysing either P-wave absence, R-R intervals or a combination of many ECG features. Ladavich and Ghoraani created a ‘rate-independent’ model that was able to identify AF using only a single cardiac cycle in order to overcome difficulties associated with detection of AF with rapid ventricular response. After feature extraction from the surface ECGs, an expectation-maximisation algorithm was used to train a model. The ultimate model had a sensitivity of 89.37% and specificity of 89.54% for detection of AF using just a single cardiac cycle.24
Longer duration analysis has also been used to provide higher accuracy in the detection of AF. He et al. used a convolutional neural network on time-frequency features (as opposed to P-wave absence or R-R intervals) to train a model for AF detection.26 The final model had an accuracy of 99.23%. Notably, the ECGs still required significant pre-processing before being classified using the convolutional neural network, which limits the potential for clinical use.
Other authors have used differing algorithms to train AF detection models. Kennedy et al. used RFs to train a model based on R-R intervals using their own database.27 They later tested this model on the MIT-BIH database. The RF model had a specificity of 98.3% and sensitivity 92.8%, which is comparatively less than seen in many of the neural-network-based approaches. However, their training database was also considerably smaller and further direct comparisons of RF and ANN models are needed to determine their comparative accuracy in AF detection.
In contrast to ANNs and RFs, Asgari et al. used an SVM to train a model for detection of AF based on wavelet transformation.28 The use of wavelet transformation obviates the need for pre-processing of P or R-waves that is required by many algorithms. However, this method still required feature extraction prior to classification with the SVM. The authors tested their model on the MIT-BIH database and found an area under the curve (AUC) of 0.995, which outperformed both a naïve Bayesian classifier and logistic regression based method.
There is less literature available on identification of ventricular arrhythmias. Mjahad et al. demonstrated that time-frequency analysis using a variety of models (logistic regression, ANN, SVM and bagging classifier) accurately identifies ventricular tachycardia (VT) and ventricular fibrillation (VF) on 12-lead ECG.25 This method required pre-processing of the ECG signal for both noise reduction and computation of time windows. On testing, all models performed similarly, with ANNs having a 98.19% accuracy for VF and 98.87% accuracy for detection of VT.
Huang et al. demonstrated the ability of ANNs to localise atrioventricular accessory pathways in patients undergoing ablation.29 Using features of delta wave polarity and R-wave duration as a proportion of the QRS complex, an ANN generated model trained on 90 cases correctly identified the site of accessory pathways in 58/60 test cases. In the two cases of misclassification, the accessory pathway was located in a contiguous region to the identified area.
The aforementioned studies have generally required significant pre-processing of the ECG signal prior to classification as well as involving models trained for a specific arrhythmia. Hannun et al. created their own test and validation database.21 This included 91,232 single lead ECGs from the Zio patch-based electrode device. These ECGs were then fed to an ANN to create a model for classifying 12 different output rhythms (10 arrhythmias, sinus rhythm and noise). Importantly, their method did not require significant feature extraction or ECG signal pre-processing. The authors validated their findings against a committee of cardiologists who had classified the test dataset. The ANN achieved an AUC of 0.91 across all rhythms. Additionally, they compared their model to that of ‘average’ cardiologists outside the adjudication committee and found that the model outperformed cardiologists across every rhythm. This work represents a significant step forward in ECG classification as it demonstrates end-to-end machine learning, where raw data is inputted and diagnostic probabilities are outputted without the use of extensive data manipulation or pre-processing.
UL methods have, thus far, had more limited application in the analysis of surface ECG. Donoso et al. used a k-means clustering algorithm to separate AF on ECG into five different types based on frequency values. However, this work is yet to be validated by examination of the clinical significance of the five different types of AF.30
AliveCor uses ML software built into their app to work in combination with an electrode band and phone or smartwatch.31 Their model is one of the first that has the ability to analyse the rhythm and diagnose AF in almost real time, as well as other ML models they have developed to identify long QT and hyperkalaemia off ECG.32,33 Twice weekly ECGs using their devices has been shown to be 3.9 times more likely to identify AF in high risk patients aged >65 years than routine monitoring.31 Additionally, recent work has examined the sensitivity of an AliveCor convolutional neural network using the Apple Watch heart rate, activity and ECG sensors as inputs compared to traditional implantable cardiac monitors for the detection of AF.3 The results suggest that the wearable monitor provides excellent sensitivity (97.5%) for detecting AF episodes lasting >1 hour but poor positive predictive value (39.9%). Given the excellent sensitivity, this strategy may help define those who would benefit from an implantable cardiac monitor post cryptogenic stroke.
Continual progress is being made in ML-based surface ECG analysis. With the advent of wearable technology, the availability of training data on which to improve models will increase and the quality of the raw data may also improve alongside the technology. However, these data will still require labelling for the implementation of SL methods, which represents a major resource barrier. External validation of the abovementioned models on larger datasets will be required before more sophisticated conclusions about the utility of such models can be made.
Intracardiac Mapping
ML is gradually emerging as a tool to improve the understanding and efficacy of ablation. Preclinical and early clinical work has focused on stratifying and classifying electrogram morphology to guide electrogram-based AF ablation.
In attempt to improve complex fractionated atrial electrogram (CFAE) based AF ablation, Schilling et al. created four classes of CFAEs based on patient data. They established classes that increased in complexity from class 0 to class 3.34 They used a variety of features in order to create their complexity classes, including time domain descriptors, phase space descriptors, wavelet based descriptors, similarity of active segments and amplitude based-descriptors. Once defined, they employed a fuzzy-decision tree (similar in methodology to a standard decision tree) to classify electrograms from 11 patients undergoing AF ablation. This algorithm had 81% accuracy for defining the CFAE class of an electrogram. The fuzzy decision tree model imparted the advantage of applying a probability that an electrogram belonged to a certain class unlike other algorithms. Duque et al. later validated and improved these CFAE classes by using a genetic algorithm (that optimised the included features) to further define the four classes.35 After the initial consolidation of the classes, they used a k-nearest neighbour SL algorithm to classify electrograms. They demonstrated 92% accuracy for classification of CFAE class and performed simulation to demonstrate that the more complex CFAE classes were associated with rotor locations in a simulated model.
Orozco-Duque et al. similarly tried to improve classification of CFAEs using four features (two time-domain morphology based and two non-linear dynamic based) to separate four classes of fractionation.36 They used a semi-supervised clustering algorithm to validate these classes on partially labelled data. They then went on to apply those classes to an unlabelled dataset and created clusters with reasonable separation. This work suggests that ML may be able to separate subtle differences in recorded electrograms and possibly provide feedback about areas most likely to result in AF termination after ablation.
McGillivray et al. used an RF classifier to identify re-entrant drivers of AF in a simulated model where the ‘true’ location of the re-entrant circuits were known.37 This model used electrogram features to assess the rhythm, predict the location and reassess the predictions until focused on the source of the driver. The model correctly identified 95.4% of drivers whether one or more were present in the simulation.
Muffoletto et al. simulated AF ablation in a 2D model of atrial tissue using three different methods – pulmonary vein isolation, fibrosis-based and rotor-based ablation.38 They were able to model the outcomes of these ablation strategies and use the outcome as labelled data for a ANN. Using different patterns in their AF simulation, the model was able to identify the successful ablation strategy in 79% of the simulations. This work serves as a proof-of-concept for ML prediction of optimal ablation strategies, but the difficulty of identifying the ‘correct’ ablation strategy amongst a number of options in vivo makes the possibility of clinical translation daunting. Computer modelling of AF is becoming more complex with the advancement of cell-level, tissue-level and organ-level models.39,40 These models have provided crucial insights into the relationships driving AF including the nature of automatic and rotational foci, the role of fibrosis and the effect of channel-types. ML may prove a useful tool in integrating the significant high-dimensional data produced by computer modelling. Possibilities include the definition of phenotypes using UL or the prediction of AF rotors based on patient-specific attributes, thus guiding ablation strategy using SL.
In an abstract presented at the American Heart Association 2018 scientific meeting, Alhusseini et al. described the use of a convolutional neural network to identify organised AF drivers that acutely terminate with ablation in persistent AF based on features extracted from spatial phase maps.41 They reported 95.2% accuracy for determining organised sites of activity that terminate AF upon ablation. These early results suggest another possibility for real-time ML-guided ablation strategies. However, the long-term efficacy of such an approach remains unclear.
ML is gradually entering the field of ablation and has the opportunity to integrate the significant volume of data generated by electroanatomical mapping systems and guide electrophysiologists to appropriate sites and methods of ablation. The use of SL in this way will face challenges given the significant data load and difficulties of creating labelled datasets, though the ultimate goal of machine-guided ablation may be achieved. However, UL methods may provide more insight into the patterns of arrhythmia seen during procedures.
Cardiac Implantable Electronic Devices
Cardiac implantable electronic devices (CIEDs) are an ideal target for ML methods given the high volume and velocity of data they produce. ML applications have allowed for risk stratification, improved arrhythmia localisation and streamlined remote monitoring which may significantly reduce the workload faced by electrophysiologists.42–44
CRT is an effective component of heart failure management in selected patients.45 Benefit is restricted to those who meet current guideline criteria based on trial data. ML models may provide a more sophisticated method of identifying those likely to respond to CRT. Kalscheur et al. used multiple SL methods on data from the Comparison of Medical Therapy, Pacing, and Defibrillation in Heart Failure (COMPANION) trial to construct a model capable of predicting outcomes with CRT.42 They employed an RF, decision tree, naïve Bayesian classifier and SVM with 48 features available in the clinical trial. They found that the RF model produced the best results, demonstrating an AUC of 0.74 for predicting outcomes. The quartile of highest risk predicted by the model had an eightfold difference in survival in comparison to the lowest risk quartile. When using traditional factors of QRS duration and morphology there was no significant association with outcomes. The features used in their model are easily clinically available. Cikes et al. employed UL for a similar purpose, seeking to identify high- and low-risk phenotypes for those likely to respond to CRT.46 Using data from the Multicenter Automatic Defibrillator Implantation Trial with Cardiac Resynchronization Therapy (MADIT-CRT) trial, they employed a k-means clustering algorithm to create groups with similar characteristics based on echocardiographic and clinical parameters. Of the four phenotypes they identified, two were associated with a better effect of CRT on heart-failure-free survival (HR 0.35 and HR 0.36 compared to non-significant). The advantage of using UL in this instance was the ability to identify the phenotypic aspects associated with CRT benefit. This is often not possible in SL owing to the nature of how the algorithms create associations, referred to as the ‘black-box’ problem.
Recognising the potential clinical difficulty of employing such models, Feeny et al. attempted to predict improved left ventricular ejection fraction with CRT using only nine features selected to optimise model performance.47 These nine variables were employed in multiple ML models (including an SVM, RF, logistic regression, an adaptive boosting algorithm and a naïve Bayesian classifier). They found that the Bayesian classifier performed best demonstrating an AUC of 0.70, which was significantly better than guideline-based prediction. To demonstrate the clinical adaptability of their approach they generated a publicly available online calculator. The above studies are clinically relevant examples of using both UL and SL to risk stratify patients and help with decision making around significant interventions. The benefits of UL as regards transparency are observed, as well as the potential difficulties around clinical adaptation of complex models.
Rosier et al. examined the potential for ML to automate monitoring of alerts from CIEDs.44 They used natural language processing to examine EHR data to determine the significance of AF alerts from CIEDs. The natural language processing algorithm was able to calculate CHA2DS2-VASc scores and anticoagulant status for each patient and thus, classify the importance of the AF alert. Their algorithm correctly stratified CHA2DS2-VASc scores of 0, 1 and ≥2 97% of the time. As a result of this, 98% of AF alerts were correctly classified with regard to their importance and the remaining misclassified alerts were overclassified, allowing for human review. This study highlights the ability of ML to act as an assistant to electrophysiologists by guiding attention to where it is needed.
In a similar vein of aiding electrophysiological intervention, Sanromán-Junquera et al. used ICD electrograms to localise exit sites during pace mapping for VT ablation.43 Using implanted RV leads as sensors, they employed multiple algorithms including ANNs, SVMs and regression methods to identify left ventricular exit sites. Their SVM model produced the best results, localising the exit site to one eighth of the heart 31.6% of the time (where a random model would produce results of 12.5%). This work serves as a proof of concept for ML based ablation localisation, especially where a 12-lead ECG of the tachycardia may not be available. However, significant improvements are required before such methods are clinically applicable.
In an application of RF models, Shakibfar et al. used only ICD data without clinical variables to predict risk of electrical storm.48 They developed 37 ICD electrogram-based features found during the four consecutive days prior to the onset of electrical storm (defined using device detection). They found that their RF model had an AUC of 0.80 for predicting electrical storm. The most relevant features were percentage of pacing and reduced daytime activity. These results are limited by the use of device-defined ventricular arrhythmias, however, they signify promise for the increasing use of the high dimensional data produced by CIEDs.
Conclusion
ML in electrophysiology is nascent. However, early work suggests the potential use that ML may have in stratification, diagnosis and therapy for arrhythmia. These methods may also affect the nature of the electrophysiologist’s role into the future with increasing data sources and methods of analysis to add to each patient’s data profile. As data derived from the use of wearable devices increases, consideration needs to be given to how these technologies will be implemented to ensure patient safety and appropriate use. In particular, the risk of over-diagnosis will need to be considered as wearable monitors become more commonplace.
Clinical Perspective
- Big data, easily collected from electronic health records and cardiac devices, allows the use of sophisticated models to generate new insights.
- Machine learning models can accurately diagnose multiple rhythms from short segments of surface electrocardiographs in almost real time.
- Optimisation of intra-cardiac mapping and implantable device analysis are areas that can significantly gain from increased machine learning integration owing to the large volume of data created in these fields.